After applying thicken all the observations of a period are mapped
to a single time point. This function will convert a datetime variable to
a character vector that reflects the period, instead of a single time point.
strftime is used to format the start and the end of the interval.
format_interval(x, start_format = "%Y-%m-%d", end_format = start_format, sep = " ", end_offset = 0, units_to_last = NULL)
Arguments
| x | A vector of class |
|---|---|
| start_format | String to format the start values of each period, to be used
in |
| end_format | String to format the end values of each period, to be used
in |
| sep | Character string that separates the |
| end_offset | Units in days if |
| units_to_last | To determine the formatting of the last value in |
Value
A character vector showing the interval.
Details
The end of the periods will be determined by the next unique value
in x. It does so without regarding the interval of x. If a specific
interval is desired, thicken and / or pad should first be
applied to create an equally spaced datetime variable.
Examples
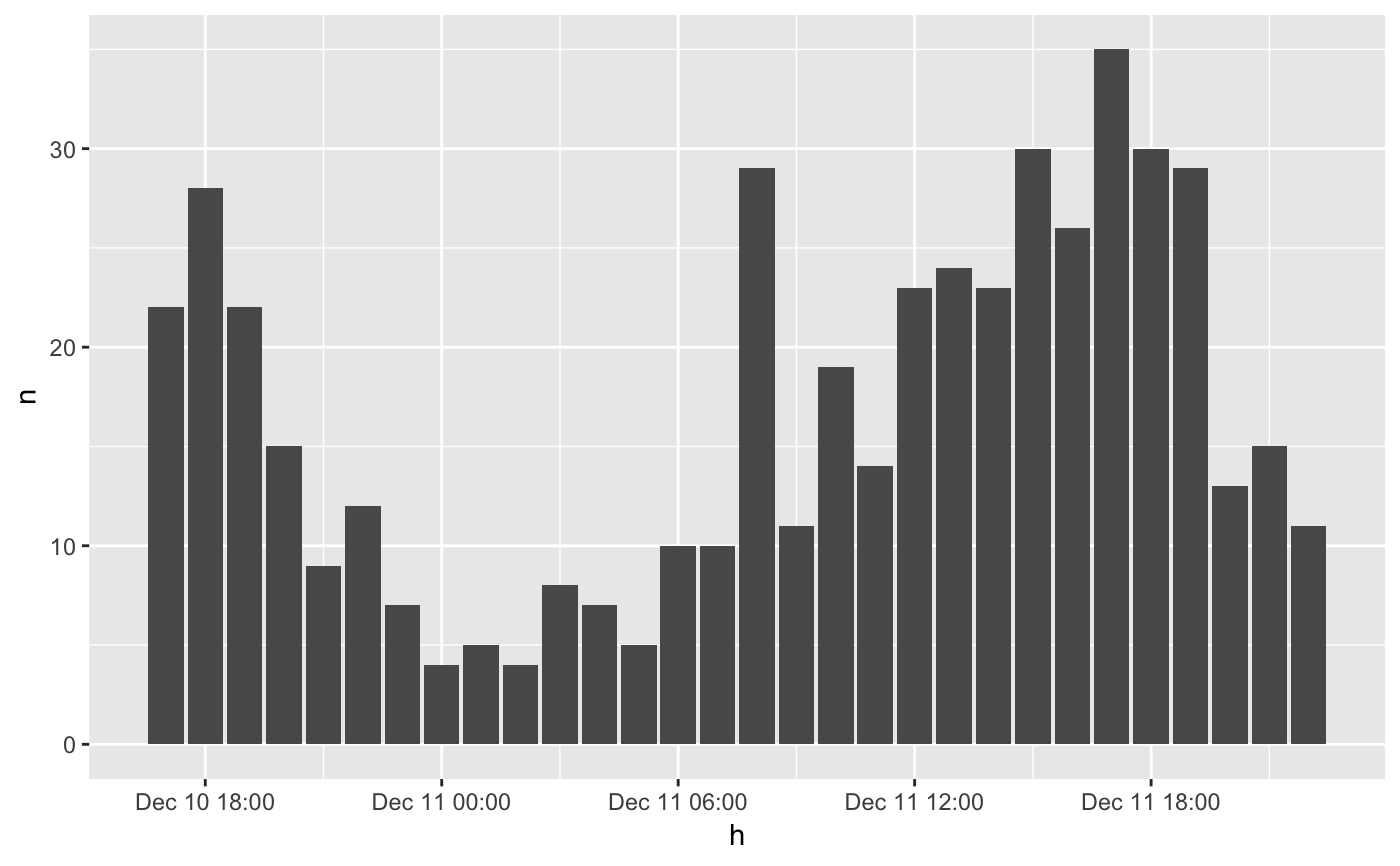
library(dplyr) library(ggplot2) plot_set <- emergency %>% head(500) %>% thicken("hour", "h") %>% count(h) # this will show the data on the full hour ggplot(plot_set, aes(h, n)) + geom_col()# adding a character to indicate the hours of the interval. plot_set %>% mutate(h_int = format_interval(h, "%H", sep = "-"))#> # A tibble: 30 x 3 #> h n h_int #> <dttm> <int> <chr> #> 1 2015-12-10 17:00:00 22 17-18 #> 2 2015-12-10 18:00:00 28 18-19 #> 3 2015-12-10 19:00:00 22 19-20 #> 4 2015-12-10 20:00:00 15 20-21 #> 5 2015-12-10 21:00:00 9 21-22 #> 6 2015-12-10 22:00:00 12 22-23 #> 7 2015-12-10 23:00:00 7 23-00 #> 8 2015-12-11 00:00:00 4 00-01 #> 9 2015-12-11 01:00:00 5 01-02 #> 10 2015-12-11 02:00:00 4 02-03 #> # … with 20 more rows